A Machine Learning-Enabled Venom Peptide Platform for Rapid Drug Discovery
Abstract
Background/Objectives: Nature has evolved millions of venom-derived peptides with diverse biological functions, a substantial fraction of which target complex membrane proteins such as G-protein-coupled receptors and ion channels. Many of these peptides are stabilized by multiple disulfide bonds, endowing them with exceptional structural stability and favorable pharmacological properties.
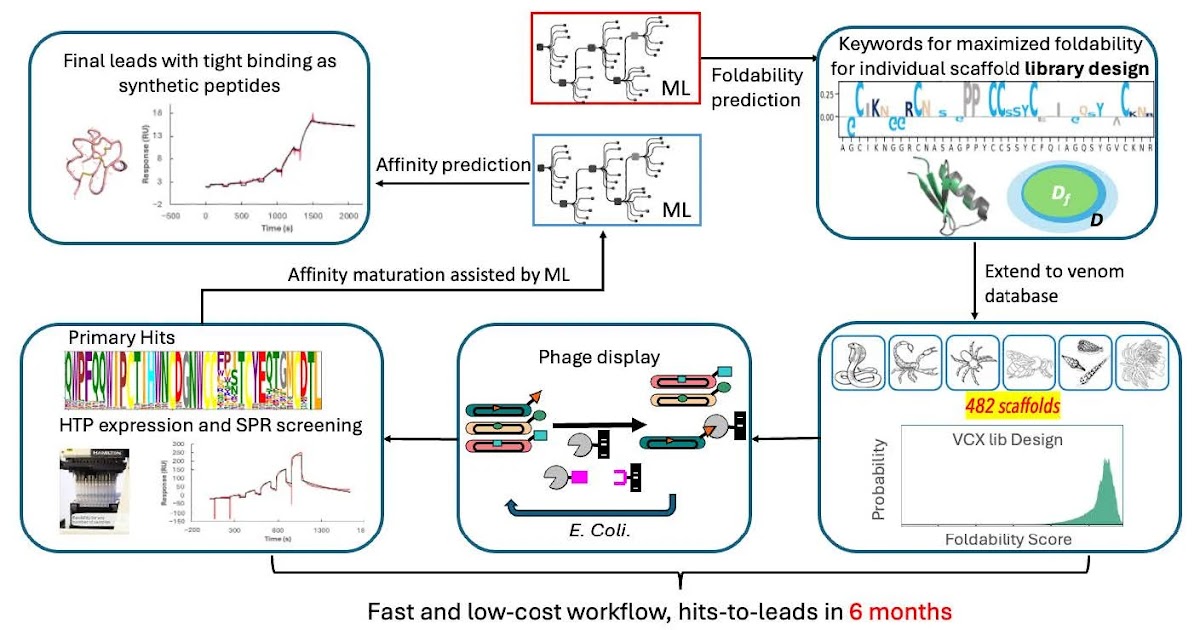
Methods: Leveraging this natural diversity, we developed a robust venom peptide therapeutics discovery system built on phage display technology and constructed a library using approximately 482 venom-derived scaffolds. The library design was guided by a machine learning (ML) model capable of predicting mutation-tolerant residues that preserve peptide foldability, maximizing structural integrity and sequence diversity.
Results: The resulting VCX library was evaluated through screening against four diverse targets (CD47, DLL3, IL33, and P2X7R), yielding strong binders for all four, a success rate of 100%. Furthermore, by integrating high-throughput recombinant expression of thioredoxin–venom fusion proteins along with ML-assisted affinity maturation, we rapidly identified potential leads for DLL3 binders.
Conclusions: This venom-based discovery platform offers significant advantages in both functionality and developability compared with conventional peptide discovery approaches. By combining natural structural diversity, ML-guided design, and recombinant expression, it enables efficient identification of “antibody-like” binders with molecular weights much smaller than those of antibodies. Consequently, it provides a powerful strategy for developing next-generation peptide therapeutics targeting challenging protein–protein interactions and complex membrane proteins.